Computation Boot Camp
Day 4 Solution
Patrick Cahan
Your final assignment:
Write a function that evaluates the groupings provided by k-means. It takes as input toyData (or a similar object), and a vector of possible k values. It returns a list of ...
- data frame of 2 columns: k, and the mean intra cluster distances
- a ggplot in which the points (samples) are colored according to the optimal value for k
- a ggplot scatter plot in which x-axis is k and the y axis is the mean intra group distance
And, apply this function to the original data set (only first 2 principle components), which you can download here:
http://cahanlab.org/intra/training/bootcampJune2016/misc/day4_assignment_data.R
What is the optimal number of clusters that your iterative kmeans approach yields?
Data sets:
- toydata
- day4_assignment_data.R (all points from day 3)
Getting started. Adjust paths as needed.
source("../../misc/utils.R")
library(ggplot2)
toyData<-utils_loadObject("../../misc/day4_toydata.R")
Here is the toy data
kmeansRes<-kmeans(toyData, 4)
kvals<-cbind(toyData, classification=kmeansRes$cluster)
ggplot(kvals, aes(x=PC1, y=PC2, colour=as.factor(classification)) ) + geom_point(pch=19, alpha=3/4, size=1) + theme_bw() +
guides(colour=guide_legend(ncol=1))
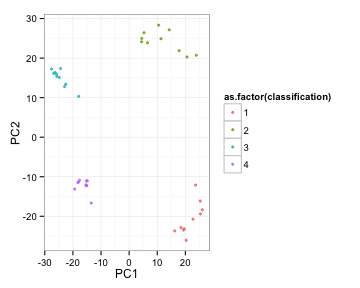
Helper function
For this assignment, I found it useful to have a little helper function
afunc<-function(adat, k){
kmeansRes<-kmeans(adat, k);
kvals<-cbind(adat, classification=kmeansRes$cluster)
ggplot(kvals, aes(x=PC1, y=PC2, colour=as.factor(classification)) ) +
geom_point(pch=19, alpha=3/4, size=1) +
theme_bw() +
guides(colour=guide_legend(ncol=2))
}
afunc(toyData, 4)
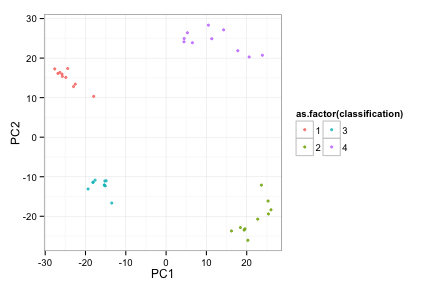
Here is the complete data
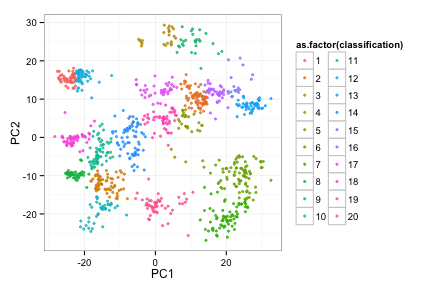
compData<-utils_loadObject("../../misc/day4_assignment_data.R")
afunc(compData, 20)
Pairwise distance calculator
eucDistV4<-function(vect1, vect2){
sum((vect1-vect2)**2)**(1/2)
}
Distances between all points
eucDistAll<-function(aMat){
myDim<-nrow(aMat);
ansMatrix<-matrix(NA, nrow=myDim, ncol=myDim);
for(i in 1:myDim){
for(j in 1:myDim){
if(j<i){
ansMatrix[i,j] <- eucDistV4(aMat[i,], aMat[j,]);
}
}
}
ansMatrix;
}
function to apply eucDistAll to all clusters returned from PCA
withinGrpDists<-function(aKvals){ # retuns a vector of the means of intra-cluster distances
grpNames<-unique(as.vector(aKvals$classification));
ans<-vector();
for(grpName in grpNames){
tmpMat<-aKvals[which(aKvals$classification==grpName),];
ans<-append(ans, mean(eucDistAll(tmpMat), na.rm=TRUE)); #Can anyone guess na.rm?
}
ans;
}
complete function, in 2 parts. don't copy and paste this
Part 1:
doRoc<-function(datMat, ks=2:20){
ans<-list(); # this is the list that will be returned upon completion
micds<-vector(); # holds means of intra-cluster distances
for(k in ks){
cat(k, " ");
tmpKres<-kmeans(datMat, k, iter.max=100);
kvals<-cbind(datMat, classification=tmpKres$cluster);
icds<- withinGrpDists(kvals);
micds<-append(micds, mean(icds, na.rm=TRUE)) # why is na.rm needed here?
}
cat("\n");
ansDF<-data.frame(k=ks, micds=micds);
bestK<-ansDF[which.min(micds),]$k;
kResFinal<-kmeans(datMat, bestK);
kvals<-cbind(datMat, classification=kResFinal$cluster);
Part 2
plot1<-ggplot(kvals, aes(x=PC1, y=PC2, colour=as.factor(classification)) ) +
geom_point(pch=19, alpha=3/4, size=1) +
theme_bw() +
guides(colour=guide_legend(ncol=2));
plot2<-ggplot(ansDF, aes(x=k, y=micds)) +
geom_point(pch=19, alpha=1, size=1) +
theme_bw();
ans[[1]]<-ansDF;
ans[[2]]<-plot1;
ans[[3]]<-plot2;
ans;
}
Test it out on the toyData
Fetch the code from http://cahanlab.org/intra/training/bootcampJune2016/misc/day4functions.R
source("../../misc/day4functions.R");
toyRes1<-doRoc(toyData)
## 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
toyRes1[[2]]
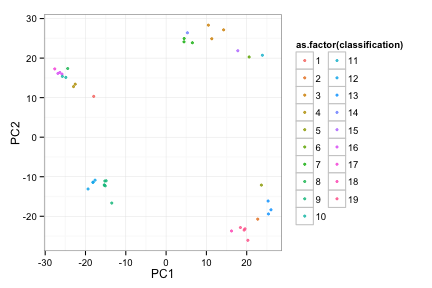
Try 1, MICDs
toyRes1[[3]]
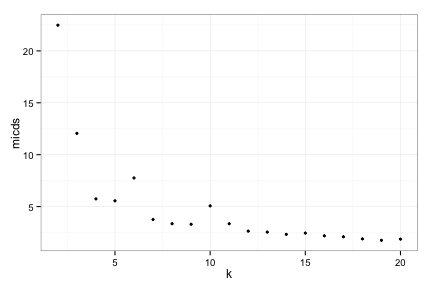
How can we make the optimal clustering jive with our intuition?
Scale MICDs by average number of samples per cluster
doRocV2<-function(datMat, ks=2:20){
nPts<-nrow(datMat); ### HERE
ans<-list(); # this is the list that will be returned upon completion
micds<-vector(); # holds means of intra-cluster distances
for(k in ks){
cat(k, " ");
tmpKres<-kmeans(datMat, k, iter.max=100);
kvals<-cbind(datMat, classification=tmpKres$cluster);
icds<- withinGrpDists(kvals);
micds<-append(micds, mean(icds, na.rm=TRUE)* k/nPts) ### HERE
}
cat("\n");
ansDF<-data.frame(k=ks, micds=micds);
bestK<-ansDF[which.min(micds),]$k;
kResFinal<-kmeans(datMat, bestK);
kvals<-cbind(datMat, classification=kResFinal$cluster);
Test it out on the toyData
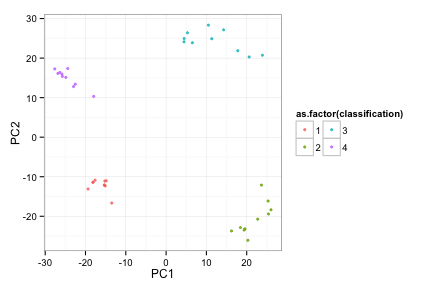
toyRes2<-doRocV2(toyData)
## 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
toyRes2[[2]]
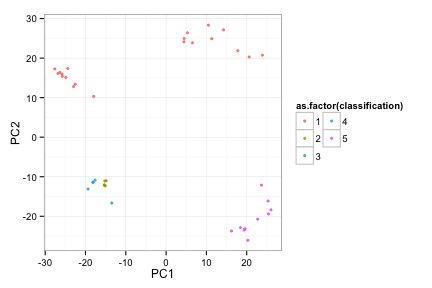
Try 2, MICDs
toyRes2[[3]]
Can we improve this further???
Singletons are ignored now, penalize them
doRocV3<-function(datMat, ks=2:20, penalize=TRUE){
nPts<-nrow(datMat);
ans<-list(); # this is the list that will be returned upon completion
micds<-vector(); # holds means of intra-cluster distances
for(k in ks){
cat(k, " ");
tmpKres<-kmeans(datMat, k, iter.max=100);
kvals<-cbind(datMat, classification=tmpKres$cluster);
icds<- wGD2(kvals); ### HERE
micds<-append(micds, mean(icds, na.rm=TRUE)* k/nPts)
}
cat("\n");
ansDF<-data.frame(k=ks, micds=micds);
bestK<-ansDF[which.min(micds),]$k;
kResFinal<-kmeans(datMat, bestK);
kvals<-cbind(datMat, classification=kResFinal$cluster);
Singletons are ignored now, penalize them
wGD2<-function(aKvals, penalize=TRUE){
grpNames<-unique(as.vector(aKvals$classification));
clusterSizes<-vector();
ans<-vector();
for(grpName in grpNames){
xi<-which(aKvals$classification==grpName)
tmpMat<-aKvals[xi,];
ans<-append(ans, mean(eucDistAll(tmpMat), na.rm=TRUE));
clusterSizes<-append(clusterSizes, length(xi)); # keep track of cluster size
}
if(penalize){
ans[which(clusterSizes==1)]<-max(ans, na.rm=TRUE); # set penalty to max micd
}
ans;
}
Test it out on the toyData
toyRes3<-doRocV3(toyData)
## 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
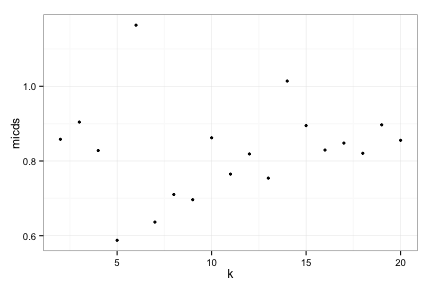
toyRes3[[2]]
Try 3, MICDs
toyRes3[[3]]
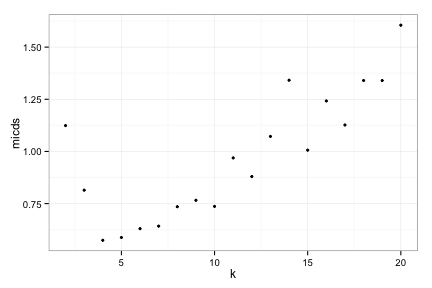
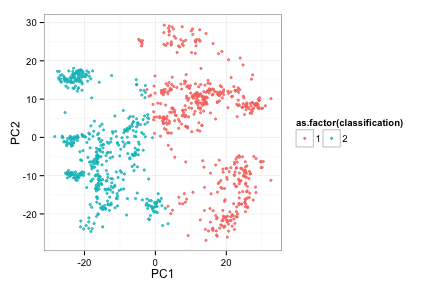
Apply to complete data set
compRes3<-doRocV4(compData) # explain how 4 is faster
## 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
compRes3[[2]]
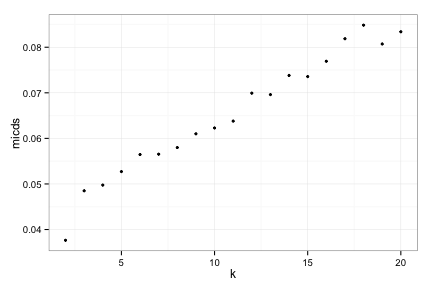
MICDs of complete data
compRes3[[3]]
Next steps:
- Explore silhouette as a metrix of clustering goodness
- Play around with the scoring to see if you can devise a more generally applicable and satisfactory metric
- Use R!